An Autosomal DNA Primer: A quick and simplified look at aspects of Autosomal testing and interpretation. Draft 1: 22-04-2024.
There are three types of Genealogical DNA tests available from Commercial Companies. They are Autosomal DNA, Y-DNA, and Mitochondrial DNA (Mt-DNA).
X-DNA data is often included with Autosomal data but is usually not useful since Mt-DNA is more effective.
The results supply information about different aspects of a person's biological heritage.
Recommended DNA Testing Companies:
(2) My Heritage. They offer Autosomal only: Click for website
(3) Ancestry DNA. They offer Autosomal testing only: Click for website
(4) GEDmatch. Autosomal and X-DNA testing only: Click for website
It is recommended that Australians purchase an Autosomal testing kit from AncestryDNA. This is the most popular test in the country. This will open up your pool of DNA matches considerably. The data from this test can then be uploaded to FTDNA, MyHeritage, and GEDmatch. The mechanics of doing so is availabe at the websites listed. Additional Information is also available on YouTube. Note that the data files can not be uploaded to Ancestry.
Y-DNA
Each human has two Haplogroups dervived from the Chromosomes of their parents, Males have an X and a Y from their Mother and Father respectivly. They get the X from their Mother, and the Y from their Father. Females normally get an X from each of their parents. Usually females don't have Y-Chromosomes, so can't take meaningful Y-DNA tests. However males down the same line of descent such as brothers, fathers, paternal uncles, or paternal grandfathers can.Besides possibly revealing DNA matches down a tester's male family line, such a test will also reveal his Y-Chromosome Haplogroup. A male will have a male haplogroup from his father and a female from his mother. The latter is defined as "the ancient group of male people from whom one's patrilineage descends". That is the sequence beginning with the chap's biological Father, Grand Father, GGfather, GGGfather, GGGGfather, etc. No Female's genetic material, or that of their male descendants is included in the results. Male Y-DNA Haplogroups have been called a "Genetic Surname". A Haplogroup is identified by a few capitalized characters, some uncapitalised, and a number, or a combination of both. For example J-M172 or E-L94 (John Aiken), or E1b1a (John Martin). World maps of the dispersion and population density of various Haplogroups are available.
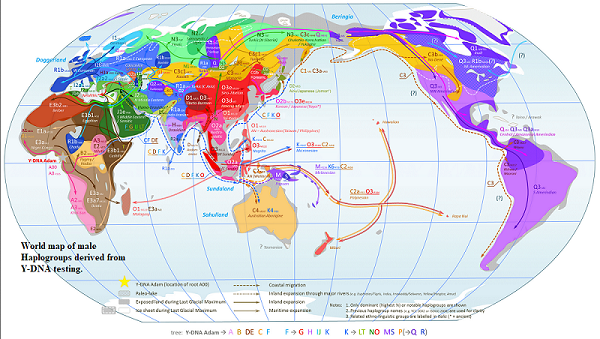
Image One, Source: https://upload.wikimedia.org/wikipedia/commons/c/ca/World_Map_of_Y-DNA_Haplogroups.png
Mitochondrial DNA
All humans have Mitochndria in their cells. They are passed down the line of decent by their mothers to their children. Both male and female can take meaningful Mt-DNA tests. These will track the DNA of the female line of their mothers, Gmothers, GGmothers etc. The tests will also return a Female alpha-numeric Haplogroup code such as U5b2b or M42a. Mitochondrial DNA tests trace people's matrilineal (mother-line) ancestry through their mitochondria, which are passed from mothers to their children. A Mt-DNA test will not reveal any information about a male line. It will however give the maternal haplogroup.
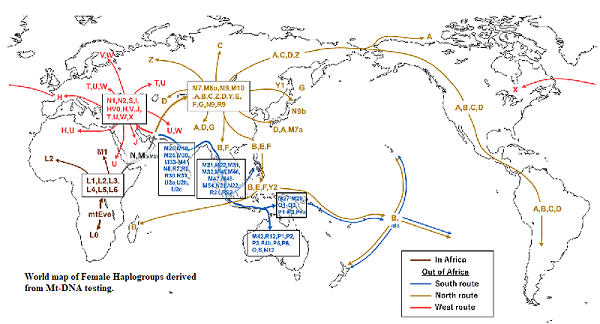
Image Two, Source: https://en.wikipedia.org/wiki/Human_mitochondrial_DNA_haplogroup
Autosomal DNA
Many of us have taken an Autosomal DNA test with Ancestry. What is Autosomal DNA? Firstly it's the ~0.3% of human DNA that is variable, and that can indicate our linage/relationships to other humans. Otherwise all humans share the same 99.5 to 99.9% of all our DNA.
Humans have Twenty Three pairs of Chromosomes as DNA, Forty Six in all in our cells. Half of each from each parent. In this case twenty two pairs of these are the Autosomes. They are numbered in the diagram from one to Twenty Two. The Twenty Third pair are the Sex Chromosomes. Labeled X and Y in the diagram below. Females have two X's. Males an X and a Y.
Interestingly while human cells normally contain Twenty Three pairs of chromosomes for a total of Forty Six, Monkeys, Chimpanzees, and Apes have Twenty-Four pairs, for a total of Fourty Eight. There is strong evidence that at some stage the Ape like pairs of Chromosome Two and Three fused together to give the human Chromosome Two. Note that the diagram below shows an X and a Y sex chromosome. It could also be two X's.
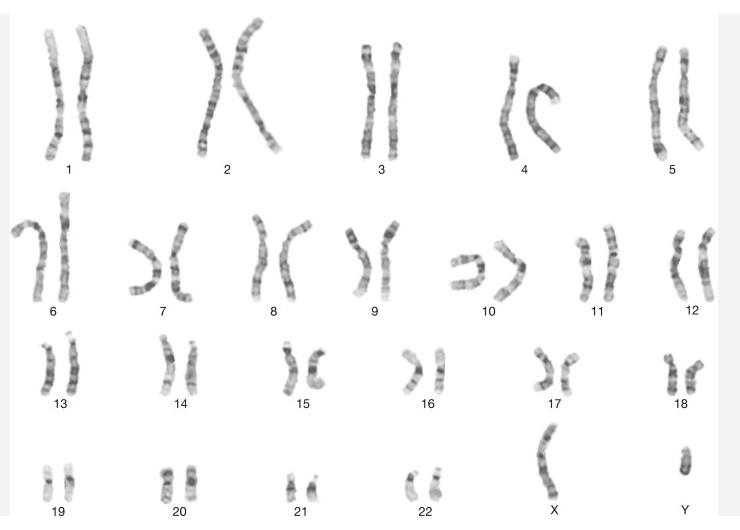
Image Three: Representation of the Twenty Three pairs of Human Cromosomes. Source sciencedirect.com
Some important definitions
Definitions of Centi-morgans.
MyfamilyTree: centiMorgan (cM) A centiMorgan (cM) is a measurement for DNA based on the likelihood of a segment to recombine from one generation to the next. A single centiMorgan is considered equivalent to a 1% (1/100) chance that a segment of DNA will crossover or recombine within one generation.AncestryDNA: A centimorgan is a unit of genetic measurement. It's what experts use to describe how much DNA and the length of specific segments of DNA you share with your relatives. These shared segments are divided up into centimorgans. The more centimorgans you share with someone, the more closely you are related.
MyHeritage: Centimorgans (cM) are units of genetic linkage between two given individuals. For example, if you share 1800 cM with an individual, that means you share around 25% of your DNA with them. A strong match will have around 200 cM or more.
23andme: A centimorgan (abbreviated as “cM”) describes the length of a piece of DNA. It is a unit of measurement. More specifically, it measures the distance between two chromosome positions. A shared DNA segment is a chunk of genetic material shared between two individuals. The length of a segment is reported in centimorgans.
GEDmatch: Centimorgan. A centimorgan is a way to measure how much DNA you share with someone else. If you receive a DNA match that shares 34 cMs across 3 segments, this indicates that you share a total of 34 centimorgans of DNA with that person. You also share a total of three DNA segments. Expressed in shorthand as cM. This is a measure of the quantity of DNA shared between two people who match on a test . A Mother/Father will share ~3600cM with each of their children. First Cousins, Great Grandparents, Great Grandchildren, and Great Aunts will share about ~800cM. Fourth Cousins will share about 35 cM, and so on. As the genetic distance between two matches increases so does the likelihood that there’s no match.
US National Human Genome Research Center: A centimorgan (abbreviated cM) is a unit of measure for the frequency of genetic recombination. One centimorgan is equal to a 1% chance that two markers on a chromosome will become separated from one another due to a recombination event during meiosis (which occurs during the formation of egg and sperm cells). On average, one centimorgan corresponds to roughly 1 million base pairs in the human genome.
Many definitions suggest that the centimorgan is a unit of distance. This is not quite true. It's really a measurement of how often something occurs. "In this case, recombination or the exchange of DNA during meiosis. The greater the distance between two genetic markers — let's say its two genes — the higher number of physical opportunities for the exchange of DNA to occur. So you have a higher frequency of recombination and a higher number of centimorgans between the genes." The unit is named after the Geneticist Thomas Hunt Morgan, a pioneer American geneticist who worked on fruit flies, a common used lab animal suitable for genetic research.
Additional Definitions:
SNP or Single Nucleotide Polymorphisms
These are very small pieces of a Chromosome that contain distinct blocks of information. Each chromosome has thousands. Equivalent pieces can be compared between people to check for a match. The information in matching SNPs is usually measured in Centimorgans.
Their so called Chromosome length or Match length expressed as cM's, is not distributed evenly down a match. Some parts have denser area's than others. There is no direct correlation between the number of SNP's and the amount of cM's.
Segment,
Defined as a block of contiguous SNP's. A section that two people share is termed a Matching Segment.
Start and End Location
When converted into numbers and letters (aka Alphanumeric form) Base Pairs, the individual markers that make up the SNPs are numbered within a Chromosome. The number of them usually run into the Millions. A segment of a chromosome can be identified by these location numbers.
Each person has about 6800 to 7200 Centimorgans of Autosomal DNA, depending how it's measured. Different companies start and finish the measuring at slightly different places on the Chromosomes.
When you send you little container of "spit" off to Ancestry in Ireland it's processed using various forms of black magic and finally a computer chip spews out a text file containing columns of numbers and letters corresponding to the location and type of the four chemical molecule present at the locations of your chromosomes. This file is available for download. The testing companies (?) use the same machine to do this processing so the data file can usually be swapped between them.
The exception is Ancestry same micro-chip notwithstanding. This seems to be a marketing decision. However their files are accepted by GEDmatch, Family TreeDNA, MyHeritageDNA etc. Until recently you could upload to their process for free and get their assessment of your DNA and a list of matches from their database. However some of them now require a small payment. GEDmatch remains free.

Image Four: DNA Match between Father/Son

Image Five: Typical AncestryDNA distant matches between two individuals.
The images above are directly from a comparison of the matches between a father and son, and between another two that could be 5th to 8th cousins. There is quite a lot of varience in the calculation of the latter because thay are genetically far apart. The next line suggests that the genetic "distance" between the two individuals is 17 Centi-Morgans. Ancestry has also calculated the distance as being less than 1% of the shared autosomal DNA. (This doesn't apply for Ethnicity Percentages by the way.) The percentage given is an alternative way of expressing the value in centiMorgans. It's found by takeing the sum of matching DNA and dividing it by 68 for Ancestry and 71.6 for to get an approximate percentage for GEDmatch and some other Companies.
Ancestry considers that the average total sum of the Autosomal DNA in centiMorgans for an individual is 6800 cM.
So for an Ancestry match between a Father and son, So the calculation becomes cM = 3444/6800 = 0.50 (rounding out to two decimal places). %cM becomes 0.50 x 100 = 50%.
The match between these distant cousins becomes: cM = 17/6800 = 0.0025. Expressed as a percentage %cM = 0.0025 x 100 = 0.25% which is recognised by Ancestry as less than 1%, that is <1%

Image Six, Table of the maximum length in Centimorgans (cM) of typical human Chrmosomes within the normal range of variability 1-22.
Sum of the cM in table for Chromosome 1 to 22 = 3587.6. Total cM for full Double Helix = 7179.2 cM
This table was created using the tools in GEDmatch and the Ancestry DATA from the father in the example shown above. It shows the spread of the centimorgans (sic) over the first 22 chromosomes. Note that the former is almost 7180 cM, 380 above the AncestryDNA assumed average of 6800. However this is well within the presumed rounding error.
Miscellaneous musings on aspects of DNA
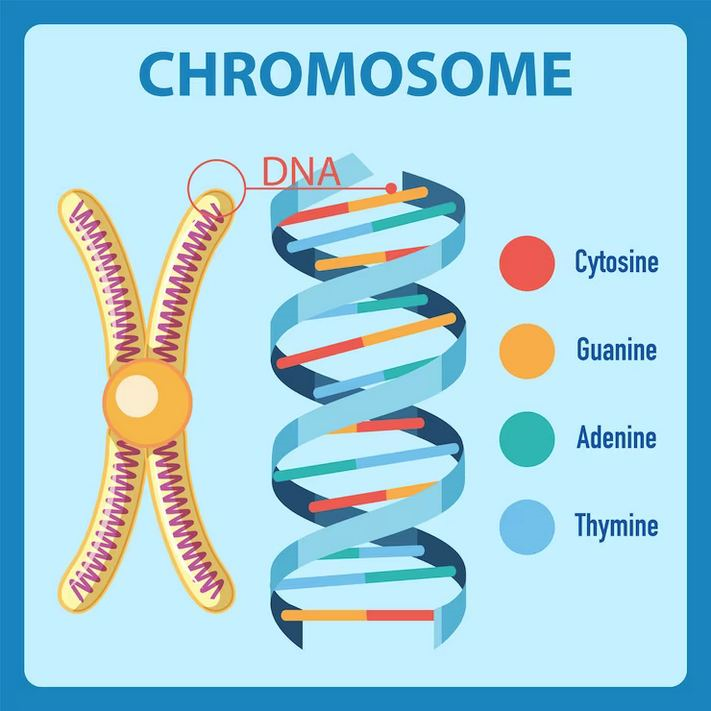
Image Seven, Connection between the Chromosome and the DNA double helix, showing the Base Pairs.Source: https://www.genome.gov/genetics-glossary/Centimorgan
Base Pair A base pair consists of two complementary DNA nucleotide bases that pair together to form a "rung of the DNA ladder". DNA is made of two linked strands that wind around each other to resemble a twisted ladder — a shape known as a double helix. Each strand has a backbone made of alternating sugar (deoxyribose) and phosphate groups. ttached to each sugar is one of four bases: adenine (A), cytosine (C), guanine (G) or thymine (T). The two strands are held together by hydrogen bonds between pairs of bases: adenine pairs with thymine, and cytosine pairs with guanine. What may not be obvious is that the Base Pairs A and T, or T and A, and the Base Pairs C and G or G and C always group together. This means that their combined lengths are exactly the same. This provides a mechanism so the connections between the outer ribbons of the DNA molecule are always the same distance apart. Hence the helical shape is maintained.
Shotgun sequencing is a laboratory technique for determining the DNA sequence of an organism’s genome. The method involves randomly breaking up the genome into small DNA fragments that are sequenced individually. A computer program looks for overlaps in the DNA sequences, using them to reassemble the fragments in their correct order to reconstitute the genome.
This is the famous "Double Helix", The colour coded Base Pairs which perform a number of important functions are called Cytosine (C), Guanine (G), Adenine (A), and Thymine (T).
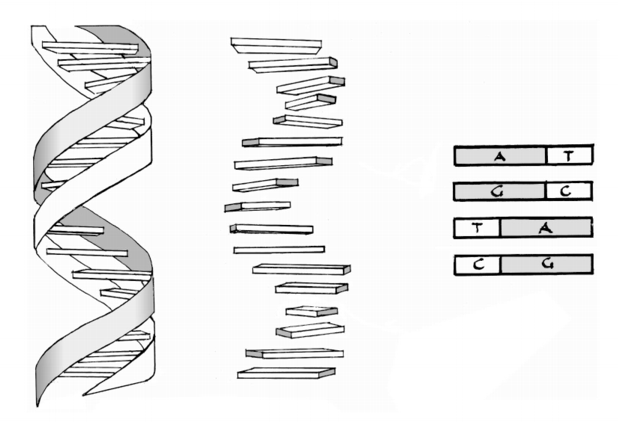
Image Eight: An alternative view of the Double Helix structure of DNA and Chromosomes emphasing the role of the Base Pairs.
Summary: Each person has over 3 Billion Bases in their DNA. Each of the latter can only be one of four types. That is a, G, C, and T.
Most of the positions of these Bases (aka a nucleotide) are the same except for a relatively small number. It's estimated that perhaps one in a thousand varies.
These are known as single nucleotide polymorphisms or SNPs (pronounced “snips”). Our genetic differences, one individual from another arise from these SNPs.
The diagram below represents the base pairs of three indivuals. The SNPs that are different are shown. All the others are the same.
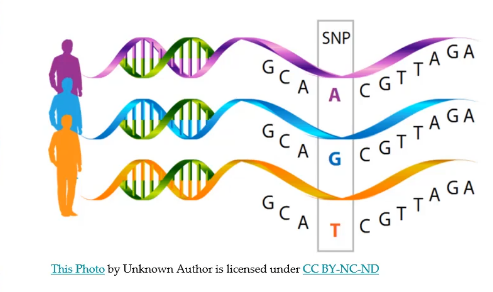
Image Nine:
Image Ten:
"Single Nucleotide Polymorphisms, frequently called SNPs ( pronounced "snips" ), are the most common type of genetic variation among people. Each SNP represents a difference in a single DNA building block, called a nucleotide. For example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA. SNPs occur normally throughout a person's DNA. They occur almost once in every 1,000 nucleotides on average, which means there are roughly 4 to 5 million SNPs in a person's genome. These variations occur in many individuals; to be classified as a SNP, a variant is found in at least 1 percent of the population. Scientists have found more than 600 million SNPs in populations around the world. Most commonly, SNPs are found in the DNA between genes. They can act as biological markers, helping scientists locate genes that are associated with disease. When SNPs occur within a gene or in a regulatory region near a gene, they may play a more direct role in disease by affecting the gene's function. Most SNPs have no effect on health or development. Some of these genetic differences, however, have proven to be very important in the study of human health. SNPs help predict an individual's response to certain drugs, susceptibility to environmental factors such as toxins, and risk of developing diseases. SNPs can also be used to track the inheritance of disease-associated genetic variants within families. Research is ongoing to identify SNPs associated with complex diseases such as heart disease, diabetes, and cancer."
Click here to return to main page
Copyright ©2023 Ray Fairall;